The CRISPR Issue: CRISPR Detects SARS-CoV-2, Treats Muscular Dystrophy in Mice & Patent Battle Gets Twisted (#8)
Plus: Transplanted stem cells produce the 'perfect' sperm, data encryption secures genetic material, and misinformation spreads on Twitter.
🧬Featured Research
CRISPR-Based Detection of SARS-CoV-2 in Under One Hour (Open Access)
A group of scientists, led by the Zhang, Gootenberg and Abudayyeh labs at MIT and Harvard, have reported a rapid method to detect SARS-CoV-2 using their Cas12-based SHERLOCK system. The test returns results in less than an hour and has a sensitivity similar to reverse-transcription–quantitative polymerase-chain-reaction (RT-qPCR) assays, the predominant method currently used to diagnose patients with COVID-19. The correspondence was published in the New England Journal of Medicine.
CRISPR Treats a Form of Molecular Dystrophy in Mice
Myotonic dystrophy type 1 (DM1) is an inherited form of muscular dystrophy, characterized by muscle wasting, weakness, clouding of the eyes, and an abnormal heartbeat. The disease is caused by “CTG microsatellite repeat expansions” in a specific gene, called DMPK (myotonic dystrophy protein kinase). Using a mouse model for DM1, a new study in Nature Biomedical Engineering injected viruses packaged with an RNA-targeting version of Cas9 to “shut down” the defective repeats, effectively reversing the symptoms of DM1 in these animals. Read the UCSD press release.
Transplanted Stem Cells Produce the Perfect Sperm (Open Access)
Farmers have been refining their breeding programs for generations, searching for the “optimal” pig, goat, or cow. To that end, artificial insemination seems to be the perfect solution: collect sperm from males with preferred traits, and introduce that sperm into the females. A new study in PNAS has shown that stem cells, transplanted into mice, pigs, goats, and cattle, can produce sperm from a genetically-preferred donor. In other words, sterile males can now produce sperm with DNA from “super sires”. For deeper coverage on this study, read the article in The Scientist.
Software Helps Design Encrypted DNA
When researchers exchange DNA—or an organism escapes from its flask—how can we figure out where the DNA came from? DNA is cheap to manufacture, so there is a pressing need to develop encryption systems for genetic materials that can link a sequence to its creator, without being counterfeited. A new study, published in ACS Synthetic Biology, reports software that can create encrypted digital signatures to incorporate into a DNA plasmid. Those “signed” plasmids “can then be ordered from a DNA synthesis company or assembled from individual DNA fragments.”
A Genetic Circuit Reduces Burden in Mammalian Cells (Open Access)
Cells have limited resources. At any given moment, thousands of processes—from DNA replication to protein production and cellular movement—are competing for those resources. When synthetic biologists engineer a cell, typically by adding DNA, those resources dwindle even more. A new study in Nature Communications partly addresses this issue in mammalian cells, demonstrating that a genetic circuit, called an “incoherent feedforward loop”, or iFFL, can be used to mitigate burden and “rescue the expression level of genes of interest despite changes in available cellular resources due to the loading effects of transgene constructs”. Read the “behind-the-paper” article by Mustafa Khammash.
🧫 Rapid-Fire Highlights
More research & reviews worth your time
Yeast were thought to have about 700 essential genes; delete them, and the cells will die (or at least be hurt pretty badly). A new study shows that 124 of the 700 or so “essential” genes in yeast are actually dispensable (Molecular Systems Biology). Open Access.
A new CRISPR homing gene drive spreads to every fly in a cage, without any observed evolution of resistance (PNAS). A separate homing gene drive, with similar results, was also published this week as a preprint (bioRxiv). Open Access.
In cell-free systems, linear DNA is chewed up and degraded. A new study explores enzymes that can protect linear DNA, and demonstrates their utility in five different bacterial cell-free systems (ACS Synthetic Biology).
How consistent can labs measure bacterial cells with optical density readings? The new iGEM Interlab study takes a closer look (Communications Biology). Open Access.
Cleveland Clinic scientists unveil a blueprint for a cheap, automated device for culturing bacteria, configured for evolution experiments. It’s so cheap, they think high school students could get one (bioRxiv). Open Access.
Preprint from Steve Quake’s group describes a “low-cost, semi-automated pipeline to extract cell-free RNA” (bioRxiv). Open Access.
Mini-review discusses methods to interface living and synthetic cells for diverse applications in bioengineering (Angewandte Chemie). Open Access.
Prime editors can make genetic substitutions, insertions, and deletions; a new study systematically evaluates prime editing off-target sites (Nucleic Acids Research). Open Access.
A new review explains how to engineer encapsulins—cage-like protein compartments—to spatially position proteins within a cell (Biotechnology and Bioengineering).
“Serratia symbiotica CWBI-2.3T is a culturable, gut-associated bacterium” found in blackflies. A new preprint brings these bacteria to the lab, showing that they can be genetically-engineered (bioRxiv). Open Access.
A new preprint uses small nanobodies to bind and recruit chromatin regulators, targeting them to specific genes in mammalian cells (bioRxiv). Open Access..
Scientists fawn over GFP, even though red fluorescent proteins have less phototoxicity and better tissue penetration. A new study uses noncanonical amino acids to red-shift fluorescent proteins (Nature Chemical Biology).
The Dunlop lab used single-cell time-lapse microscopy to measure gene expression and growth of E. coli over many generations; fifteen fluorescent reporters were used to track genes related to the stress response, delineating how stochastic fluctuations between cells can determine which cells live, and which die (bioRxiv). Open Access.
📰 #SynBio in the News
There was a painful abundance of misinformation about synthetic biology that circulated on Twitter this week (leading to at least one account suspension), especially surrounding the idea that SARS-CoV-2 was engineered in a lab. This is a claim for which there is zero credible evidence. Don’t @ me.
Natalie Ma, from Felix Biotechnology, was interviewed for this week’s Titus Talks podcast.
OpenTrons, the small robotics company, has started a coronavirus testing lab in New York City that can return results in 24-48 hours.
Synthetic biologists at Codagenix have created an attenuated form of the new coronavirus that “has a ‘virtual brake pedal’ inside, causing it to replicate much less quickly.” Antonio Regalado reported for the MIT Technology Review.
The CRISPR patent battle will likely continue after the Patent Trial and Appeal Board finds that “the Broad Institute has ‘priority’ in its already granted patents for uses of the original CRISPR system in eukaryotic cells,” according to an article for Science by Jon Cohen.
New research shows that "Viking” is not a term based on heredity; it was a job title. Andrew Curry reported for Science.
Startup company Wildtype is taking pre-orders for its lab-grown salmon, according to a new story on Tech Crunch.
A fascinating Nature news feature that I missed last week: how Parabon Nanolabs is “using DNA to sketch the faces of criminals".
An op-ed for bioeconomy.xyz explained the benefits, and costs, of a career in biotech and academia.
The ASBMB (American Society for Biochemistry and Molecular Biology) published a featured story on metabolic engineering and limonene, featuring Claudia Vickers.
An op-ed in Foreign Policy warns of the dangers of synthetic biology using, in my opinion, a plethora of recycled ideas. Scientists on Twitter fired back.
🐦Science Threads on Twitter
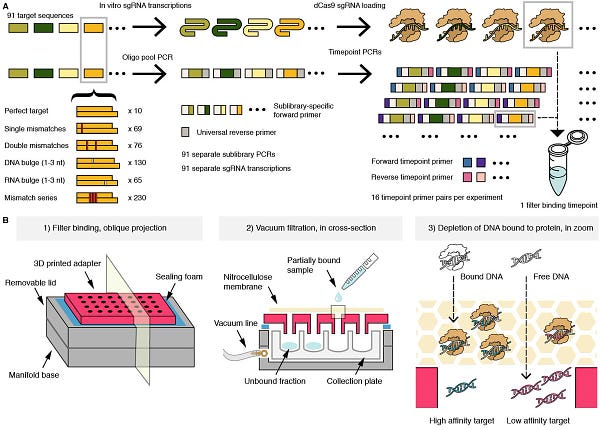
The Twitter highlight this week features a preprint from the Doudna & Greenleaf labs. In this work, led by Evan Boyle, the authors performed “62,444 quantitative binding and cleavage assays on 35,047 on- and off-target DNA sequences across 90 Cas9 ribonucleoproteins (RNPs) loaded with distinct guide RNAs”. Wow! Check out the Twitter thread. 👇

Thanks for reading This Week in Synthetic Biology, part of Bioeconomy.XYZ. As always, I welcome your feedback via direct Twitter message or comment. If you enjoy this newsletter, please share it with a friend.
A version of these newsletters is posted on bioeconomy.xyz and my website, hiniko.io. Reach me with tips and feedback via Twitter direct message at @NikoMcCarty.